SPECIES FOR WHICH ORTHOLOGOUS EXONS / CDS ARE AVAILABLE
| EUARCHONTOGLIRES | ||
| Homo_sapiens | Human | v01 |
| Pan_troglodytes | Chimp | v01 |
| Gorilla_gorilla | Gorilla | v05 |
| Pongo_pygmaeus | Orangutan | v03 |
| Nomascus_leucogenys |
Gibbon |
v07 |
| Macaca_mulatta | Macaque | v01 |
| Papio_anubis |
Olive baboon |
v09 |
| Chlorocebus_sabaeus |
Vervet |
v09 |
| Callithrix_jacchus | Marmoset | v06 |
| Tarsius_syrichta | Tarsier | v05 |
| Otolemur_garnettii | Bushbaby | v02 |
| Microcebus_murinus | Mouse lemur | v02 |
| Tupaia_belangeri | Tree shrew | v02 |
| Mus_musculus | Mouse | v01 |
| Rattus_norvegicus | Rat | v01 |
| Dipodomys_ordii | Kangaroo rat | v05 |
| Cavia_porcellus | Guinea pig | v02 |
| Ictidomys_tridecemlineatus | Ground squirrel | v02 |
| Oryctolagus_cuniculus | Rabbit | v01 |
| Ochotona_princeps | Pika | v02 |
| LAURASIATHERIA | ||
| Bos_taurus | Cow | v01 |
| Ovis_aries |
Sheep |
v09 |
| Tursiops_truncatus | Dolphin | v05 |
| Sus_scrofa | Pig | v06 |
| Vicugna_pacos | Alpaca | v05 |
| Equus_caballus | Horse | v03 |
| Canis_familiaris | Dog | v01 |
| Ailuropoda_melanoleuca |
Panda |
v07 |
| Mustela_putorius_furo |
Ferret |
v08 |
| Felis_catus | Cat | v02 |
| Myotis_lucifugus | Microbat | v02 |
| Pteropus_vampyrus | Megabat | v05 |
| Erinaceus_europaeus | Hedgehog | v02 |
| Sorex_araneus | Shrew | v02 |
| AFROTHERIA | ||
| Loxodonta_africana | Elephant | v01 |
| Procavia_capensis | Hyrax | v05 |
| Echinops_telfairi | Tenrec | v01 |
| XENARTHRA | ||
| Dasypus_novemcinctus | Armadillo | v01 |
| Choloepus_hoffmanni | Sloth | v05 |
| MARSUPIALIA | ||
| Monodelphis_domestica | Opossum | v01 |
| Macropus_eugenii | Wallaby | v06 |
| Sarcophilus_harrisii |
Tasmanian devil |
v07 |
| MONOTREMATA | ||
| Ornithorhynchus_anatinus | Platypus | v02 |


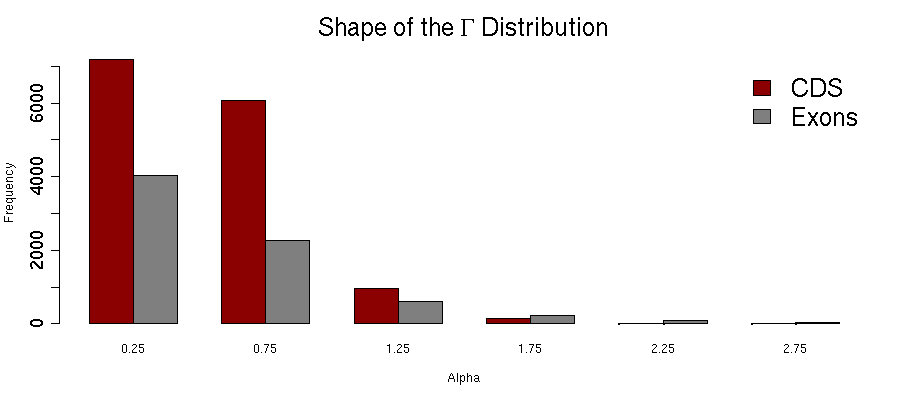
a SHAPE OF THE G DISTRIBUTION
The substitution rate heterogeneity among sites of the exon / CDS alignment is described by the Γ distribution.PERCENTAGE OF G+C ON THIRD CODON POSITIONS
Lower (respectively higher) α values correspond to strong (respectively weak) heterogeneity. If α > 1, the substitution pattern among sites is rather homogeneous.
Request guidelines: In the [0-5] range of values, type alpha < 1 to query markers with stronger among-sites variability, and alpha > 1 for alignments with more evenly distributed variability.
Reference: Yang Z., 1996.
Among-site rate variation and its impact on phylogenetic analyses.
Trends in Ecology and Evolution 11 (9) : 367-372.
This parameter is a descriptor of the degree of base composition heterogeneity.
It is more contrasted on third codon positions than on the whole exon / CDS.
Request guidelines: In the [20-95] range of values, type %GC3 < 40 to query A+T rich markers, 40 < %GC3 < 60 for more equilibrated markers, and %GC3 > 60 for G+C rich markers.





